Research Areas
Transcription Factor Circuitries in Ovarian Cancer
Characterizing the transcription factors (TFs) deregulated during tumorigenesis has provided key insights into disease etiology, disease origins and therapeutic targeting for many tumor types; however, in ovarian cancer, researchers know very little about the TF networks responsible for maintaining cell state and viability. The Lawrenson Laboratory has amassed a large compendium of enhancer landscapes (using H3K27ac ChIP-seq) and transcriptomes (using RNA-seq) for ovarian cancer histological subtypes and precursor cells, and we leverage these data to identify master regulators driving the development of ovarian cancers.
One TF with a clear role in ovarian cancer is PAX8. PAX8 is highly expressed in the majority of high-grade serous and clear cell ovarian cancers, and can also be detected in some of the rarer histotypes. Variants at this locus are associated with mucinous ovarian cancer (Kelemen, Lawrenson et al. Nat Genet. 2015 Aug;47(8):888-897. https://www.nature.com/articles/ng.3336). In collaboration with colleagues at the University of Cambridge in the U.K., the Lawrenson Lab has shown that PAX8 target genes are enriched at serous ovarian cancer risk loci (Kar et al. Br J Cancer. 2017 Feb 14;116(4):524-535. https://www.nature.com/articles/bjc2016426). Moreover, recent analyses show that PAX8 is both a target of and mediator of noncoding somatic variation in ovarian cancer (Corona et al. Preprint. https://www.biorxiv.org/content/10.1101/537886v1).
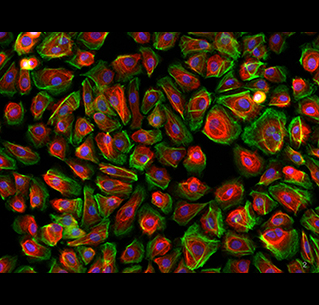
Immunofluorescent staining of monolayer cultured cells
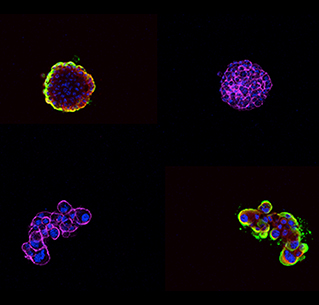
Immunofluorescent staining of three-dimensional ovarian cancer cell models
Characterizing the lncRNA Landscape of Ovarian Cancer
Very little is known about the role of long noncoding RNAs (lncRNAs) in epithelial ovarian cancer (EOC) development. Research in the Lawrenson Laboratory uses RNA-sequencing technologies to catalogue the lncRNA transcriptome of ovarian cancers and their precursor tissues: specifically, ovarian surface epithelial cells and fallopian tube secretory cells for the most common subtype (high-grade serous). A current focus of the Lawrenson Lab is a specific lncRNA—UCA1—that is associated with poor outcomes in EOC. In vitro studies indicate that this long noncoding RNA induces metabolic reprogramming in ovarian cancer cells and perturbs a key pathway involved in development and cancer. Current investigations in the Lawrenson Lab are exploring the mechanisms by which UCA1 promotes ovarian cancer development and whether UCA1-regulated pathways represent molecular vulnerabilities in UCA1-driven cancers that can be targeted therapeutically.
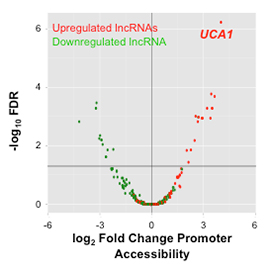
| Integrating RNAseq expression data with epigenetic biofeatures to identify lncRNAs differentially expressed in ovarian cancer cells compared to normal ovarian epithelial cells. |
Elucidating the Function of Risk and Somatic Variants Associated with Ovarian Cancer
Genome-wide association studies have identified a plethora of genetic variants that influence a person’s risk of developing complex traits, including cancer. The overwhelming majority of these variants lie in noncoding DNA. More recently, whole-genome sequencing studies have catalogued on average 10,000 variants per ovarian tumor, the majority of which are outside of coding exons. These analyses highlight a central role for gene regulation in cancer risk and somatic development, and so it has been necessary to develop novel approaches to the functional analysis of these polymorphisms: first, to identify the regulatory biofeature coinciding with the variant and, second, to identify and characterize the target gene or genes. The Lawrenson Lab uses in vitro models, genome editing, ChIP-seq and RNA-seq to identify and test the function of novel variants and their target genes.
Identifying Novel Therapeutic Approaches for Women's Cancers
Three-dimensional cell culture models and cell culture systems developed from biologically relevant tissues represent unique resources for profiling active pathways in disease and responses to chemotherapeutic agents or targeted therapies. The Lawrenson Laboratory has generated 3-D models of EOC precursor tissues and shown that 3-D cell culture models of ovarian cancer cell lines can recreate the histological diversity of ovarian cancers. Moreover, 3-D ovarian cancer cultures also tend to be more chemoresistant, indicating that 3-D culture systems are more stringent drug development platforms. The Lawrenson Laboratory is now using these novel 3-D model systems to test novel therapeutic strategies for the treatment of ovarian cancer.
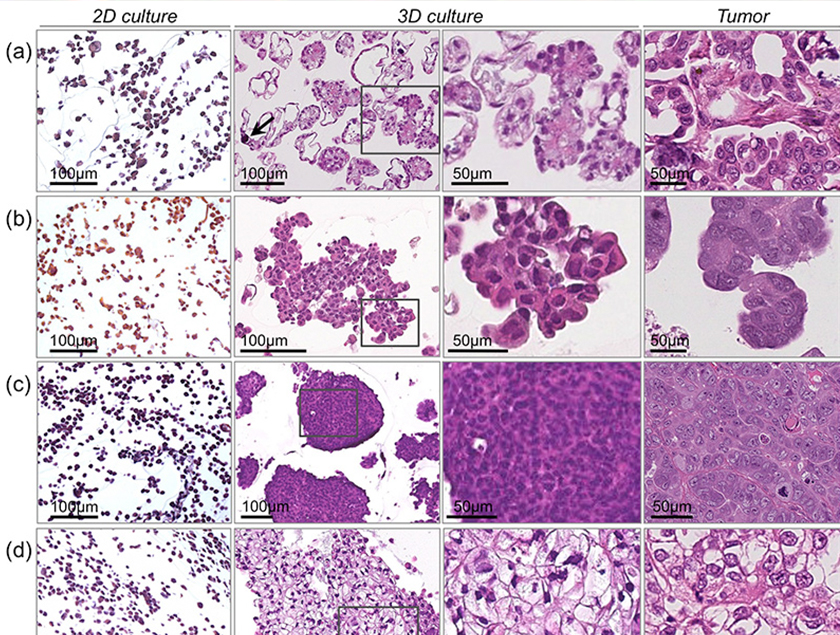
Three-dimensional cultured ovarian cancer cells more closely mimic the histology of human ovarian cancer histological subtypes: (a) low-grade serous ovarian cancer, (b) high-grade serous ovarian cancer, (c) poorly differentiated ovarian cancer (d) clear cell ovarian cancer. From Lee et al. Lab Invest. 2013 May;93(5):528-542. |
Discovering Molecular Drivers and Clinical Biomarkers for Endometriosis
Endometriosis is characterized by endometrial glands and stroma outside of the uterine cavity, causing chronic pain, dysmenorrhea and infertility. Endometriosis is present in up to 10 percent of reproductive-aged women. True prevalence of the disease in the general population is likely grossly underestimated, as many women may be asymptomatic, and the associated symptoms are variable and not specific to this disease (e.g., pelvic pain). Consequently, a definitive diagnosis usually requires surgery.1 In addition to pain and infertility, endometriosis is associated with increased risk of ovarian cancer, particularly the clear cell and endometrioid ovarian cancer subtypes (the endometriosis-associated ovarian cancers, EnOCs).2-6
The economic burden of endometriosis is substantial,7 with the direct costs per patient in the U.S. reaching over $12,000 annually. Indirect costs are also large, primarily due to loss of work productivity as a result of pain and illness. The total financial burden of endometriosis in the U.S. is now thought to exceed $75 billion dollars annually.8 In spite of this, many basic questions about endometriosis etiology remain unanswered. For example, endometriosis is divided into four stages (I to IV) to describe the extent of the adhesions and implants observed. However, it is not clear whether the different stages represent different parts of the same disease continuum or distinct endpoints. Endometriosis can also be described according to disease site: ovarian endometriosis (endometrioma), deep infiltrating disease, and peritoneal lesions. Deep infiltrating disease is the most aggressive subtype, with cancer-like invasive behavior and frequent mutations in known cancer driver genes. Somewhat paradoxically, however, this subtype of endometriosis is not associated with increased ovarian cancer risk, whereas many studies have now shown that ovarian endometriosis is associated with increased risk.9,10
To address these crucial questions, the Lawrenson Laboratory has established the Biological and Epidemiological Markers of Endometriosis Study at Cedars-Sinai, working in close collaboration with the surgeons in the Minimally Invasive Gynecologic Surgery Center. The study is collecting tissues and blood from affected patients and has ongoing studies to understand cellular and transcriptional heterogeneity in endometriosis and identify novel circulating biomarkers.
- Hickey M, et al. Endometriosis. BMJ. 2014 Mar 19;348:g1752.
- Kohl Schwartz AS, et al. Endometriosis, especially mild disease: a risk factor for miscarriages. Fertil Steril. 2017;108(5):806-814.e2.
- Sainz de la Cuesta R, et al. Histologic transformation of benign endometriosis to early epithelial ovarian cancer. Gynecol Oncol. 1996;60(2):238-244.
- Lee AW, et al. Evidence of a genetic link between endometriosis and ovarian cancer. Fertil Steril. 2016;105(1):35-3.e1.
- Pearce CL, et al. Association between endometriosis and risk of histological subtypes of ovarian cancer: a pooled analysis of case-control studies. Lancet Oncol. 2012;13(4):385-394.
- Lu Y, et al. Shared genetics underlying epidemiological association between endometriosis and ovarian cancer. Hum Mol Genet. 2015;24(20):5955-5964.
- Soliman AM, et al. The direct and indirect costs associated with endometriosis: a systematic literature review. Hum Reprod. 2016;31(4):712-722.
- Simoens S, et al. The burden of endometriosis: costs and quality of life of women with endometriosis and treated in referral centres. Hum Reprod. 2012;27(5):1292-1299.
- Anglesio MS, et al. Cancer-associated mutations in endometriosis without cancer. N Engl J Med. 2017;376(19):1835-1848.
- Saavalainen L, et al. Risk of gynecologic cancer according to the type of endometriosis. Obstet Gynecol. 2018;131(6):1095-1102.
Contact the Lawrenson Lab
8700 Beverly Blvd.
Davis Building, Third Floor
Los Angeles, CA 90048
